PCA
Principal component analysis is a commonly used dimensionality reduction method, which can transform many highly correlated features into a few features that are independent or unrelated to each other.
For example, the data with 6 samples, 4 features
x1 |
x2 |
x3 |
x4 |
|
1 |
7 |
26 |
6 |
60 |
2 |
1 |
29 |
15 |
52 |
3 |
11 |
56 |
8 |
20 |
4 |
11 |
31 |
8 |
47 |
5 |
7 |
52 |
6 |
33 |
6 |
11 |
55 |
9 |
22 |
Perform dimension reduction on x1,x2,x3,x4
A |
|
1 |
[[7,26,6,60],[1,29,15,52],[11,56,8,20],[11,31,8,47],[7,52,6,33],[11,55,9,22],[3,71,17,6],[1,31,22,44],[2,54,18,22],[21,47,4,26],[1,40,23,34],[11,66,9,12],[10,68,8,12]] |
2 |
=pca(A1) |
3 |
=A2(2).((~)/sum(A2(2))) |
4 |
=A3.(cum(~)) |
5 |
=pca@r(A1,2) |
A1 The rows of the matrix correspond to the samples and the columns correspond to the features
A2 PCA is performed on A1, return information for dimension reduction

A2(1) Returns the average of each column
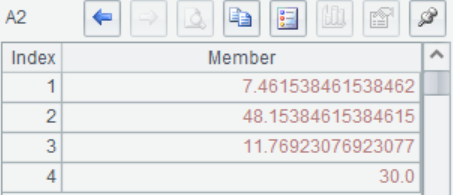
A2(2) The principal component variance, which is the eigenvalue corresponding to each eigenvector, is arranged from largest to smallest.

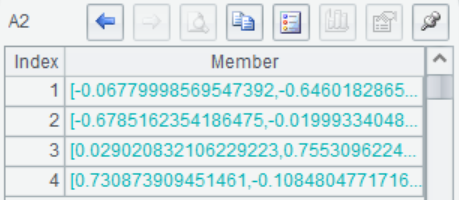
A1(3) Return to the principal component coefficient which is the eigenvector matrix of the covariance matrix
A3 The information contribution rate of four eigenvalues

A4 Calculate the cumulative contribution rate, it can be seen that the sum of the first two features has reached more than 97%, so the first two principal components can be taken.

A5 Dimension reduction is performed on A1, n value is 2, and the result of dimension reduction is returned

SPL Official Website 👉 https://www.scudata.com
SPL Feedback and Help 👉 https://www.reddit.com/r/esProc_SPL
SPL Learning Material 👉 https://c.scudata.com
SPL Source Code and Package 👉 https://github.com/SPLWare/esProc
Discord 👉 https://discord.gg/cFTcUNs7
Youtube 👉 https://www.youtube.com/@esProc_SPL


